4 A simple example
4.1 Data import
The data in question is part of a survey, where patients who came to a health center for having their blood pressure measured were asked about lifestyle factors, including physical activity and nutrition. For the current example, we only have a small subset of \(n=100\) subjects, for whom we have age, sex, systolic and diastolic blood pressure as well their answer to the question “Do you regularly add salt to your meal after it has been served?”. We are interested in how the risk factors sex and “extra salt” are related to the blood pressure variables.
Looking at the file saltadd.txt in a text editor (e.g. notepad on Windows), we notice that
- the first row of the file contains variable names, and
- the columns of the data seem to be separated by tabulators (tabs).
We therefore can read the file with the command
In other words, we command R to take the file named saltadd.txt in the sub-directory Data of the current working directory,
and read it in
- using a tabulator5 as separator between fields in the same row,
- using the first row to set the column names,
- converting non-numerical variables into factors.
The resulting data frame is then stored as object salt_ht in the current working environment, where we can process it as we see fit.
Alternatively, you can import the data file via opening the drop-down menu Import Data in the Environment tab and selecting From Text (base). After selecting the data file via the GUI, this opens a dialogue where you can set the appropriate options for the data import, including the name of the resulting data frame, the column separator, and conversion of text columns to factors, just as above. RStudio will translate your input into the corresponding call to function read.delim, which is a variant of read.table with different default settings (see ?read.delim for details).
4.2 Analysis
4.2.1 Descriptives
Let’s start with a simple summary of the whole data, to familiarize ourselves with it, and to do some general quality control:
> summary(salt_ht)
ID sex sbp dbp saltadd age
Min. :1006 female:55 Min. : 80.0 Min. : 55.00 no :37 Min. :26.00
1st Qu.:2879 male :45 1st Qu.:121.0 1st Qu.: 80.00 yes :43 1st Qu.:39.75
Median :5237 Median :148.5 Median : 96.00 NA's:20 Median :50.00
Mean :5227 Mean :154.3 Mean : 98.51 Mean :48.71
3rd Qu.:7309 3rd Qu.:184.0 3rd Qu.:116.25 3rd Qu.:58.00
Max. :9962 Max. :238.0 Max. :158.00 Max. :71.00 These seem overall reasonable values, though the blood pressure of the subjects do tend towards high and even very high values (e.g. systolic blood pressures above 200 points).
Note that the salt variable actually has 20 missing values (coded as NA in R); any comparison involving this variable will by default only use the 80 subjects where the variable has non-missing values.
4.2.2 Blood pressure by salt intake
Let’s look at the distribution of the systolic blood pressure by salt intake. For a small data set like this, side-by-side boxplots work well for comparison, so we use the function boxplot with the formula interface we have seen before:
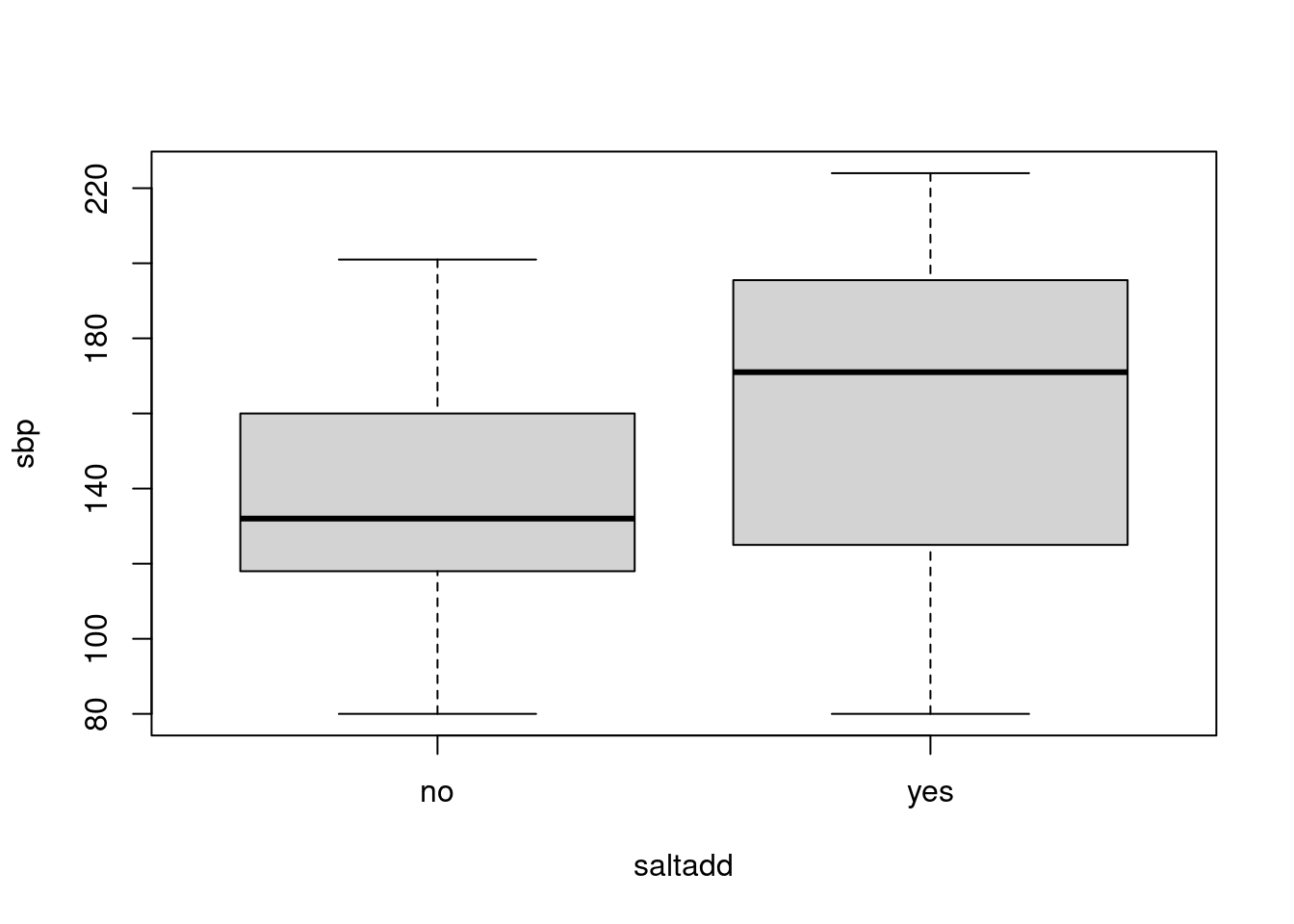
It appears that the median blood pressure is higher in those who add extra salt, though both groups also show considerable variability, as indicated by the box heights (interquartile ranges).
We also want to report the actual mean- and median blood pressures for both groups separately. We can do this using the subset function we have introduced earlier:
> salt_ht_yes <- subset(salt_ht, saltadd == "yes")
> salt_ht_no <- subset(salt_ht, saltadd == "no")
> summary(salt_ht_yes$sbp)
Min. 1st Qu. Median Mean 3rd Qu. Max.
80.0 125.0 171.0 163.0 195.5 224.0
> summary(salt_ht_no$sbp)
Min. 1st Qu. Median Mean 3rd Qu. Max.
80.0 118.0 132.0 137.4 160.0 201.0 Note that this is not an efficient or recommended way of doing sub-group analysis in R, but it is what works with our still very limited R vocabulary. Per-group data processing will be discussed in more detail in Section 6.
The question remains whether the difference in blood pressure we see is small or large in relation to the sampling variation in the data. We can test the null hypothesis that the underlying true mean systolic blood pressure is the same in both groups using the function t.test with the now already customary formula notation:
> t.test(sbp ~ saltadd, data = salt_ht)
Welch Two Sample t-test
data: sbp by saltadd
t = -3.3088, df = 76.733, p-value = 0.001429
alternative hypothesis: true difference in means between group no and group yes is not equal to 0
95 percent confidence interval:
-40.95533 -10.17981
sample estimates:
mean in group no mean in group yes
137.4324 163.0000 The result is a Welch t-test (i.e. not assuming equal variances in both groups). R reports the name of the test, the test statistic, degrees of freedom and the resulting p-value plus the alternative hypothesis (here indicating that this is a two-sided p-value). We conclude that at the usual significance level of \(\alpha=0.05\), there is indeed a statistically significant difference between the mean systolic blood pressures in the two underlying populations. Additionally, the output also lists a 95% confidence interval for the mean difference as well as the group means.
Exercises:
- How can we calculate the standard deviation of the systolic blood pressure in both groups?
- How can we run a Student t-test using the function
t.test? - Repeat the analysis, but for the diastolic blood pressure; add an informative title to the boxplot.
4.2.3 Salt intake by sex
We also want to look a the relationship between sex and salt intake. As both of these are categorical, we just need to look at the corresponding cross-tabulation. This can be done via the function table simply by specifying two arguments instead of one as before:
At first glance, it seems that the salt intake does indeed differ by sex. As before, we can use the function proportions to convert the asbolute frequencies to proportions / relative frequencies:
> tab <- table(salt_ht$sex, salt_ht$saltadd)
> proportions(tab)
no yes
female 0.1625 0.3500
male 0.3000 0.1875Note that for a two-by-two table like here, we have different possibilities for calculating proportions: we can report the proportion of the total, like above, or we can calculate row- or column-proportions (i.e. split by salt intake within sex, or split within salt intake by sex). If we want one of the latter, we can specify the margin as second argument to proportions, with 1 indicating rows and 2 indicating columns:
> proportions(tab, margin = 1)
no yes
female 0.3170732 0.6829268
male 0.6153846 0.3846154
> proportions(tab, margin = 2)
no yes
female 0.3513514 0.6511628
male 0.6486486 0.3488372So we have e.g. 68% of women who usually add salt, as compared to only 38% of men.
We can use the function chisq.test to test the null hypothesis that there is no association between the rows (i.e. sex) and the columns (i.e. salt intake) of this frequency table:
> chisq.test(tab)
Pearson's Chi-squared test with Yates' continuity correction
data: tab
X-squared = 6.0053, df = 1, p-value = 0.01426Note that the results of this test look quite similar to what we have seen from the t-test above: name of the test, test statistic and degrees of freedom, p-value (and it appears that men and women do differ significantly in their intake of salt in this example, at least at the conventional \(\alpha=0.05\)).
4.2.4 Save results
As final touch, we want to save the data frame we have imported for further processing. This can be done via the function save which takes any number of objects as arguments, and saves them to the binary data file specified via argument file:
A quick check with dir will show that we now have the new file saltadd.RData in the folder Data. The content of this file (i.e. the single data frame salt_ht) can be loaded into any R session either via the function load at the console, by clicking on the file name in the Files tab, or via the open folder-icon in the top left of the Environment tab.
4.3 Turning it into a script
A proper statistical analysis consists of two elements, the original raw data and the code used to generate the reported output - everything else is just messing around with data. Specifically, a result file is not an analysis, but only the output from an analysis.
This means we are not done yet - we still have to turn our interactive session into a proper R script. We have already looked at the mechanics of this process in Section 3.3.1: highlight all relevant commands in the History tab and click on To Source to copy them to the Source pane, either to a freshly opened new script file or just into the currently open text file.
Here, let’s assume that we have saved the commands we just copied to from the History tab to a script file called BasicExample.R. This is still a very crude affair: hard to read, may well continue incorrect commands (typos), and completely lacks context. This cannot stand, and we should perform at a minimum the following four steps:
Clean up the code: delete repeated commands, incorrect commands, help calls, explorations that do not contribute to the final analysis etc. (short is good).
Add a comment header that very briefly explains what the script does, who has written it, and roughly when the script was first written. Whoever opens the file should not have to scroll to see this minimal information.
Arrange the code in smaller, logical units separated by empty lines; each unit should correspond to one step in the overall analysis, similar to how we use paragraphs to structure ideas in a paper.
Add comments throughout, explaining what you do and why. You don’t have to go overboard, but it should provide sufficient context to understand your reasoning. A reasonable ambition level is that you yourself should understand what is going on six months after you have last touched the script (harder than it sounds!).
For our minimal example in this section, a reasonable script could look like this:
# BasicExample.R
#
# Example code for course Introduction to R (2958)
# Module 1: A scripted data analysis example
#
# Alexander.Ploner@ki.se 2024-03-08
# Load the data; note explicit path to file
salt_ht <- read.table("~/Rcourse/Data/saltadd.txt", header = TRUE,
sep = "\t", stringsAsFactors=TRUE)
# Descriptives: quality control
summary(salt_ht)
# Q1: systolic blood pressure by saltadd - descriptives
boxplot(sbp ~ saltadd, data = salt_ht)
salt_ht_yes <- subset(salt_ht, saltadd == "yes")
salt_ht_no <- subset(salt_ht, saltadd == "no")
summary(salt_ht_yes$sbp)
summary(salt_ht_no$sbp)
# Q1: inference
t.test(sbp ~ saltadd, data = salt_ht)
# Q2: salt intake by sex - descriptives
tab <- table(salt_ht$sex, salt_ht$saltadd)
tab
# Proportion saltadders per sex
proportions(tab, margin=1)
# Q2: inference
chisq.test(tab)
# Save the imported data frame; note - same explicit path
# as for original text file
save(salt_ht, file="~/Rcourse/Data/saltadd.RData")4.4 Exporting results
Simply running an analysis script will recreate the original analysis results in RStudio, with numerical output in the console and graphical output in the Plots tab. Alternatively, R scripts can be compiled to create a document that contains both the original R commands and the output they generate. At the simplest level, the resulting document corresponds to a log file in other statistical programs like Stata or SAS, which serves as a record of running a script on a specific data set.
In order to generate such an output file, simply load the script into the RStudio Source Pane, and click on the notebook icon  (alternatively, press the hotkey combination Ctrl-Shift-K). This will open a small dialogue with a choice of three output formats, HTML, PDF and MS Word. Clicking on Compile will generate an output file with the same basename as the script file and the file extension corresponding to the chosen output format.
(alternatively, press the hotkey combination Ctrl-Shift-K). This will open a small dialogue with a choice of three output formats, HTML, PDF and MS Word. Clicking on Compile will generate an output file with the same basename as the script file and the file extension corresponding to the chosen output format.
Figure 4.1 below shows what this looks like: in the output file, the comments and code from the original script (boxplot, subset, summary) are combined with the output they produce, both graphical (the boxplots) and numerical (the six-number summaries). Together, this is a self-contained record of both the analysis steps and their results.
If we compile the script to a file format that is a bit easier to edit than HTML, say the MS Word format, the resulting output file can also serve as the starting point for a report on the analysis and its results.
Note that this is just a starting point for exporting results from R/RStudio. We will see that it is easy to add R functions to a script that will format results in an attractive manner in the compiled document (Sections 8.5 and 13), and easy to write comments that appear as formatted text (Section 14). Together, this will allow us to turn a simple R script into a very reasonable draft report with just the click of a button.
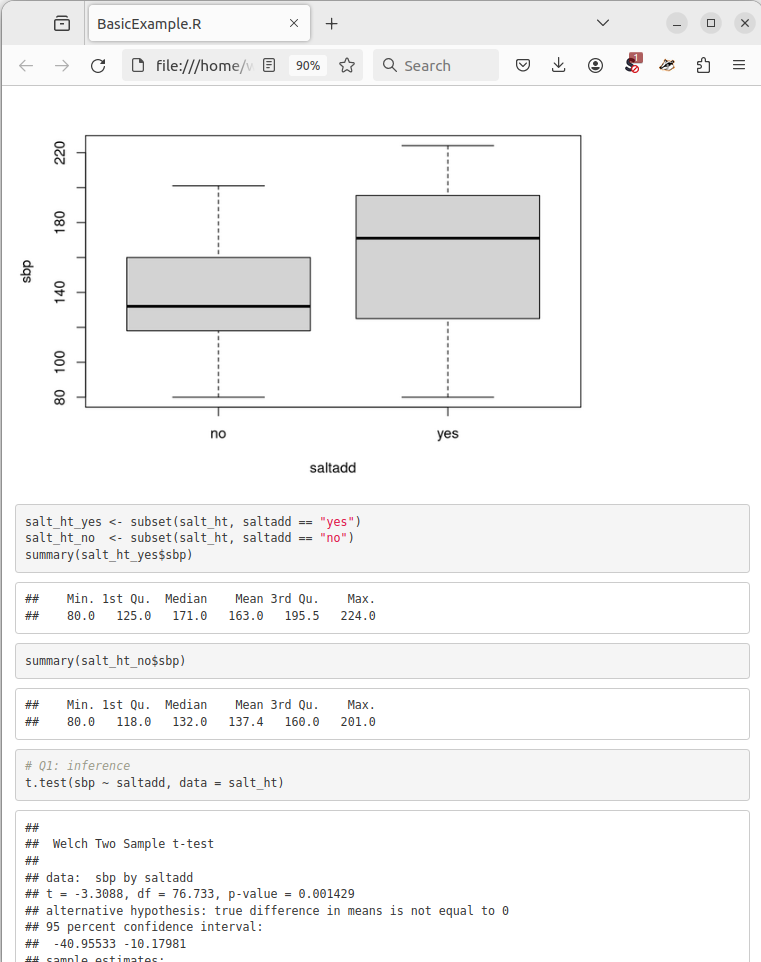
Figure 4.1: Extract from BasicExample.html, compiled from script BasicExample.R
Note that
\trepresents a special character (ASCII code 9) used to indicate indentation and separation (e.g. in data files as in this example); the combination\tis not a string of two characters, but represents the single (unprintable) tab character, see e.g. https://en.wikipedia.org/wiki/Tab_key#Tab_characters↩︎